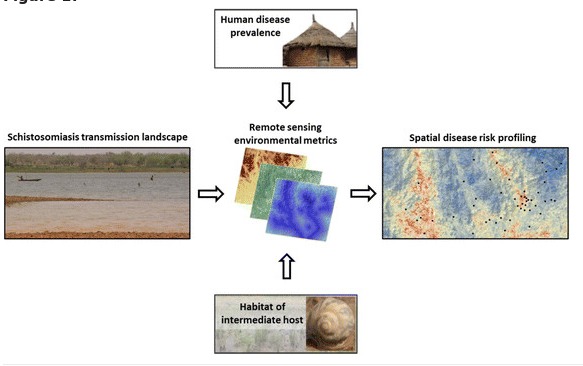
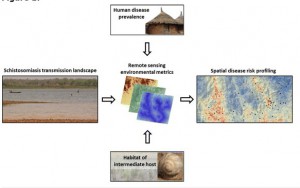
 The review article lead by Yvonne Walz is published online first. Schistosomiasis is a water-based disease that affects an estimated 250 million people, mainly in sub-Saharan Africa. The transmission of schistosomiasis is spatially and temporally restricted to freshwater bodies that contain schistosome cercariae released from specific snails that act as intermediate hosts. Our objective was to assess the contribution of remote sensing applications and to identify remaining challenges in its optimal application for schistosomiasis risk profiling in order to support public health authorities to better target control interventions.
The review article lead by Yvonne Walz is published online first. Schistosomiasis is a water-based disease that affects an estimated 250 million people, mainly in sub-Saharan Africa. The transmission of schistosomiasis is spatially and temporally restricted to freshwater bodies that contain schistosome cercariae released from specific snails that act as intermediate hosts. Our objective was to assess the contribution of remote sensing applications and to identify remaining challenges in its optimal application for schistosomiasis risk profiling in order to support public health authorities to better target control interventions.
We reviewed the literature (i) to deepen our understanding of the ecology and the epidemiology of schistosomiasis, placing particular emphasis on remote sensing; and (ii) to fill an identified gap, namely interdisciplinary research that bridges different strands of scientific inquiry to enhance spatially explicit risk profiling. As a first step, we reviewed key factors that govern schistosomiasis risk. Secondly, we examined remote sensing data and variables that have been used for risk profiling of schistosomiasis. Thirdly, the linkage between the ecological consequence of environmental conditions and the respective measure of remote sensing data were synthesised.
We found that the potential of remote sensing data for spatial risk profiling of schistosomiasis is – in principle – far greater than explored thus far. Importantly though, the application of remote sensing data requires a tailored approach that must be optimised by selecting specific remote sensing variables, considering the appropriate scale of observation and modelling within ecozones. Interestingly, prior studies that linked prevalence of Schistosoma infection to remotely sensed data did not reflect that there is a spatial gap between the parasite and intermediate host snail habitats where disease transmission occurs, and the location (community or school) where prevalence measures are usually derived from.
Our findings imply that the potential of remote sensing data for risk profiling of schistosomiasis and other neglected tropical diseases has yet to be fully exploited.